Output directory structure¶
Author: Menachem Sklarz
Table of Contents
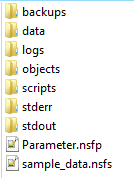
The main directory structure¶
The directories are elaborated on below.
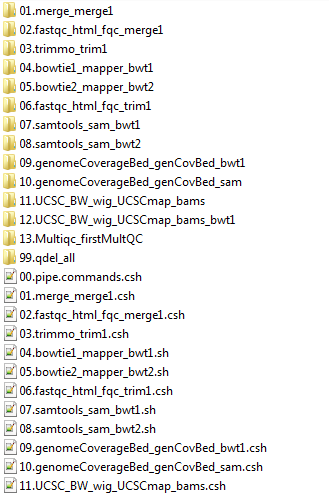
The scripts directory¶
- Executing
bash 00.workflow.commands.shwill execute the entire workflow. - The scripts beginning
01.Import…etc. execute entire steps. - The actual scripts running each step per sample or on the entire project are contained in the equivalent directories
01.Import…etc. - The scripts are numbered by execution order (see
00.workflow.commands.sh)
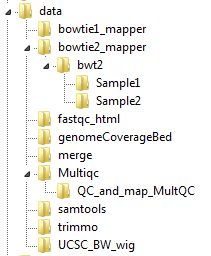
The data directory¶
In the data directory, the analysis outputs are organized by module, by module instance and by sample.
Below is the data directory for the example, showing the tree organization for the bowtie2_mapper and Multiqc modules.
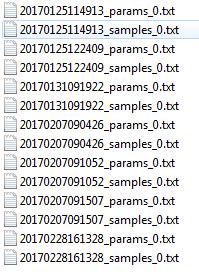
The backup directory¶
The backup directory contains a history of workflow sample and parameter files.
The logs directory¶
The logs directory contains various logging files:
- version_list. A list of all the versions of the workflow with equivalent comments
- file_registration. A list of files produced, including md5 signatures, and the script and workflow version that produced them
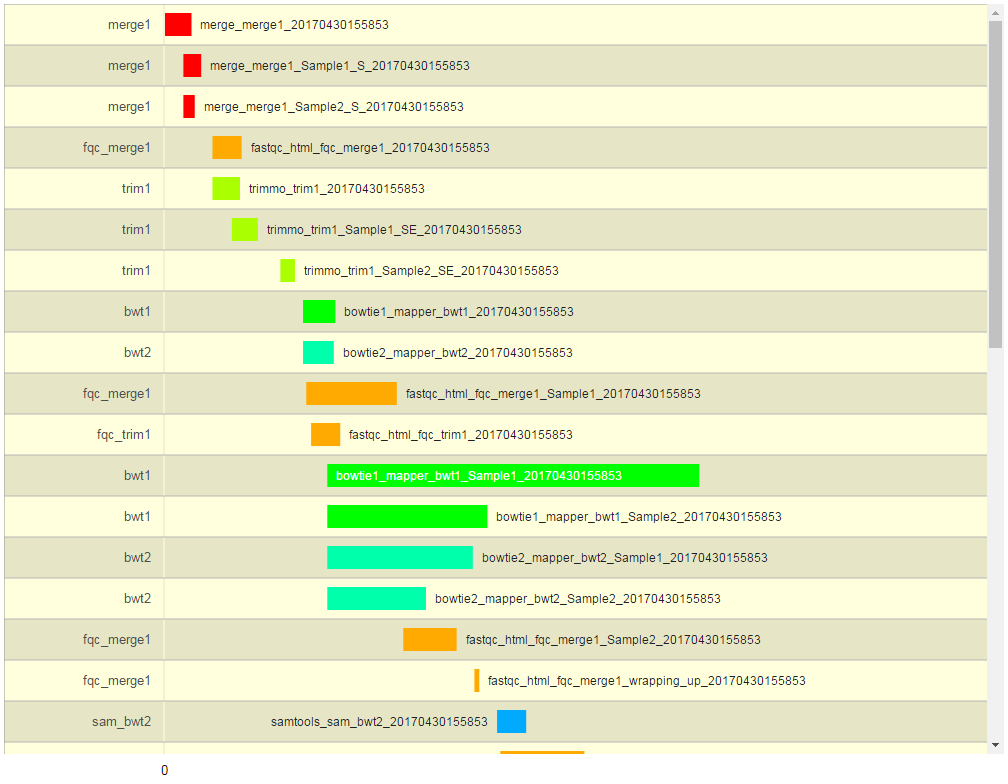
log_file_plotter.R. An R script for producing a plot of the execution times. (Run with Rscript and receives a single argument – a log file to plot)log_<workflow_ID>.txt. Log of the execution times of the script per workflow version ID.log_<workflow_ID>.txt.html. Graphical representation of the progress of the WF execution, as produced by thelog_file_plotter.Rscript (see figure below)
The stderr and stdout directories¶
- The stderr and stdout directories store the script standard error and outputs, respectively.
- These are stored in files containing the module name, module instance, sample name, workflow ID and cluster job ID.
The objects directory¶
The objects directory contains various files describing the workflow:
pipeline_graph.html: An SVG diagram of the workflow.diagrammer.R: an R script for producing a DiagrammeR diagram of the workflow.pipedata.json: A JSON file containing all the workflow data, for uploading to JSON compliant databases etc.workflow_graph.htmlis the output from executingRscript diagrammer.R.