NeatSeq-Flow Tutorial Workflow¶
| Author: | Menachem Sklarz and Liron Levin |
|---|---|
| Affiliation: | Bioinformatics Core Facility |
| Organization: | National Institute of Biotechnology in the Negev, Ben Gurion University. |
Module categories
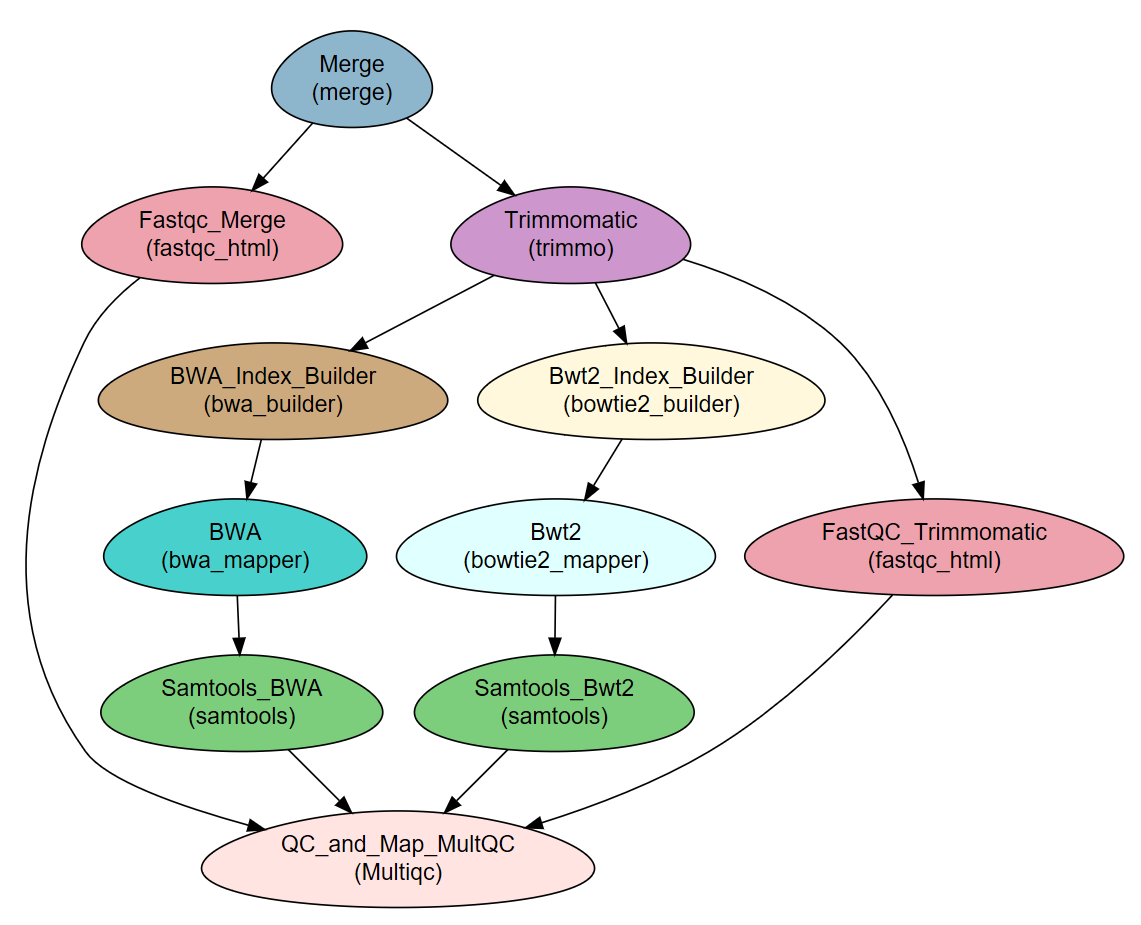
This tutorial describes how to create and execute the workflow described in the NeatSeq-Flow manuscript (Article on BioRXiv).
See NeatSeq-Flow Tutorial for detailed instructions for quick installation of the tutorial workflow with conda.
The example workflow receives FASTQ files and the sequenced genome of a bacteria. It then performs:
- Quality testing and trimming of the raw sequence reads (paired- or single-end).
- Alignment (“mapping”) of the reads to a reference genome using two different programs.
- Sorting the samples’ BAM files as final results.
- Creation of a report on reads and mapping quality.
Steps¶
| Step | Module | Program |
|---|---|---|
| Merge | merge | |
| Fastqc_Merge | fastqc_html | fastqc |
| Trimmomatic | trimmo | trimmomatic |
| FastQC_Trimmomatic | fastqc_html | fastqc |
| BWA_Index_Builder | bwa_builder | bwa |
| BWA | bwa_mapper | bwa |
| Bwt2_Index_Builder | bowtie2_builder | bowtie2 |
| Bwt2 | bowtie2_mapper | bowtie2 |
| Samtools_BWA | samtools | samtools |
| Samtools_Bwt2 | samtools | samtools |
| QC_and_Map_MultQC | Multiqc | MultiQC |
Required data¶
This WF requires samples with fastq file(s) (paired or single) and a reference genome in fasta format.
Programs required¶
- fastqc
- trimmomatic
- multiqc
- samtools=1.3
- BWA
- bowtie2
Example of Sample File¶
Title Example_WF_From_the_manuscript
#Type Path
Nucleotide /path/to/Reference_genome.fasta
#SampleID Type Path
Sample1 Forward /path/to/Sample1.F.fastq.gz
Sample1 Reverse /path/to/Sample1.R.fastq.gz
Sample2 Forward /path/to/Sample2.F.fastq.gz
Sample2 Reverse /path/to/Sample2.R.fastq.gz
Sample3 Forward /path/to/Sample3.F.fastq.gz
Sample3 Reverse /path/to/Sample3.R.fastq.gz